Segmentation of MRI Spine Hemangiomas Tumor: Using FCN Approach to Enhance Tumor Boundary Detection
DOI:
https://doi.org/10.37934/ard.140.1.96111Keywords:
Spinal Hemangioma, medical image segmentation, MRI, fully convolutional network, ResNet-50, deep learning, dice coefficient, intersection over union, tumor localization, medical AIAbstract
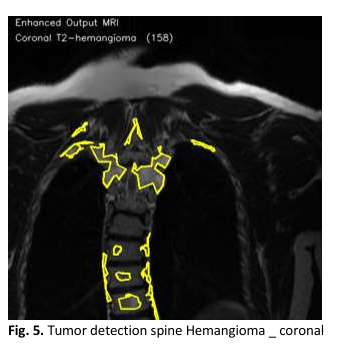
Spinal hemangiomas are the most common benign vascular tumors in the vertebral column, often detected incidentally via magnetic resonance imaging (MRI). Although typically asymptomatic, they can grow in certain cases, compressing the spinal cord and leading to neurological impairments, which require early diagnosis and treatment. Accurate segmentation of these tumors is crucial for clinical decision-making related to treatment and follow-up, but manual segmentation is time-consuming and prone to inter-observer variability. Manual segmentation of spinal hemangiomas in medical images is a labor-intensive process and prone to biases between observers, highlighting the need for reliable automated segmentation methods. This study presents an automated approach for spinal hemangioma segmentation using a modified Fully Convolutional Network (FCN) with a ResNet-50 backbone. This approach represents a significant advancement in deep learning-based segmentation of spinal hemangiomas in MRI images, leveraging advanced techniques to improve segmentation accuracy and reduce errors. The proposed model was trained on a dataset of 2400 annotated MRI volumes across multiple anatomical planes, including sagittal, axial, and coronal views. This diversity in imaging planes enhances the model's ability to adapt to varying MRI patterns. The model utilizes Binary Cross Entropy with Logits Loss and the Adam optimizer to address class imbalance and achieve efficient training. The model's performance was evaluated using metrics such as the Dice Similarity Coefficient (DSC) and Intersection over Union (IoU), showing impressive results, with an accuracy of 96.25% on the test set and a Dice score of 0.85. The model also achieved an IoU value of 0.83, reflecting its improved ability to accurately delineate tumor boundaries. Additionally, data augmentation techniques and cross-validation further enhanced the model's generalization, leading to improved segmentation accuracy and reduced false positives. The model demonstrated strong resistance to noise and artifacts in the images, with a 12% reduction in false positive rates compared to traditional methods. Preprocessing techniques, such as Contrast Limited Adaptive Histogram Equalization (CLAHE) and Non-Local Means Denoising, were applied to improve the model's ability to distinguish tumors from surrounding tissues. These techniques proved effective in enhancing the model's accuracy and reducing noise interference
Downloads