In-Silico Modeling of Pattern Recognition Receptor EFR and Molecular Interaction with Pathogen Associated Molecular Pattern elf18
Keywords:
PAMP, EFR, MD Simulation, elf18Abstract
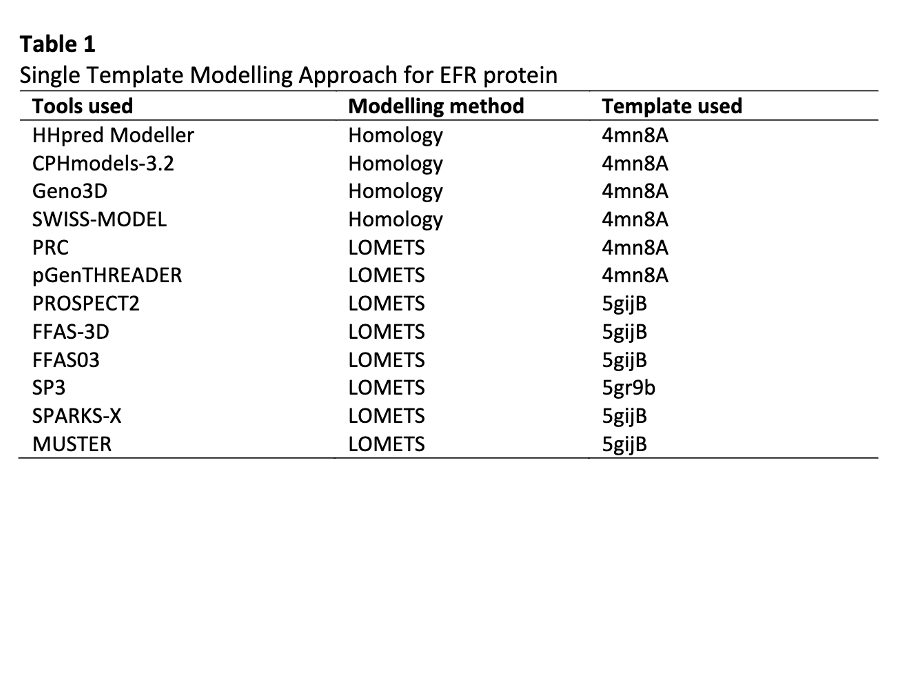
Plants depend entirely on pattern-triggered immunity (PTI) system to protect them from various pathogenic bacteria. It is activated by pathogen-associated molecular pattern (PAMP) of the host plant by pattern recognition receptor (PRR) with coreceptor. Elongation Factor Receptor (EFR) is one of the PRR used to protect against Brassica species disease. Although research on transgenic approach have been carried out to analyze the EFR protein, but the full ectodomain interactions of EFR with PAMP elf18 protein and co-receptor Brassinosteroid Insensitive 1-associated receptor kinase (BAK1) through in-silico has not been accomplished yet. The purpose of this study was to determine the interaction of EFR protein with elf18 protein through in-silico analysis. In this study, PRR EFR and PAMP elf18 was constructed by homology modeling using HHpred Modeler, followed by docking and molecular dynamics (MD) simulations of EFR and elf18 protein using Z-Dock and GROMACS 5.0.4 respectively. Modelling results showed that multiple template modeling (MTM) generated best models compared to single template modeling (STM) due to their best quality of the protein structure obtained by HHpred Modeler generate best-validation results of 71.123 ERRAT, 95.67% Verify3D and 92.8% in favoured region of the Ramachandran Plot. Docking results showed that the complex interaction of BAK1 and elf18 binds at leucine-rich-repeat (LRR) EFR (LRR 1-8 and LRR 12-14). After 50ns MD Simulation, the results showed that the docked complexes have significant reduction of H-bonds. For EFR-elf18-BAK1 (normal) complex, 20 hydrogen bonds were sustained compared to EFR-elf18-BAK1 (mutated) complex that only sustained 16 hydrogen bonds, proved that the mutated protein have less interaction after simulation. This study may contribute significantly towards understanding the early event of Pattern Triggered Immunity mechanism of EFR-elf18-BAK1 protein complex.
Downloads