PoMSA: An Efficient and Precise Position-Based Multiple Sequence Alignment Technique
Keywords:
Multiple Sequence Alignment, computational biology, bioinformatics, Heuristic algorithmAbstract
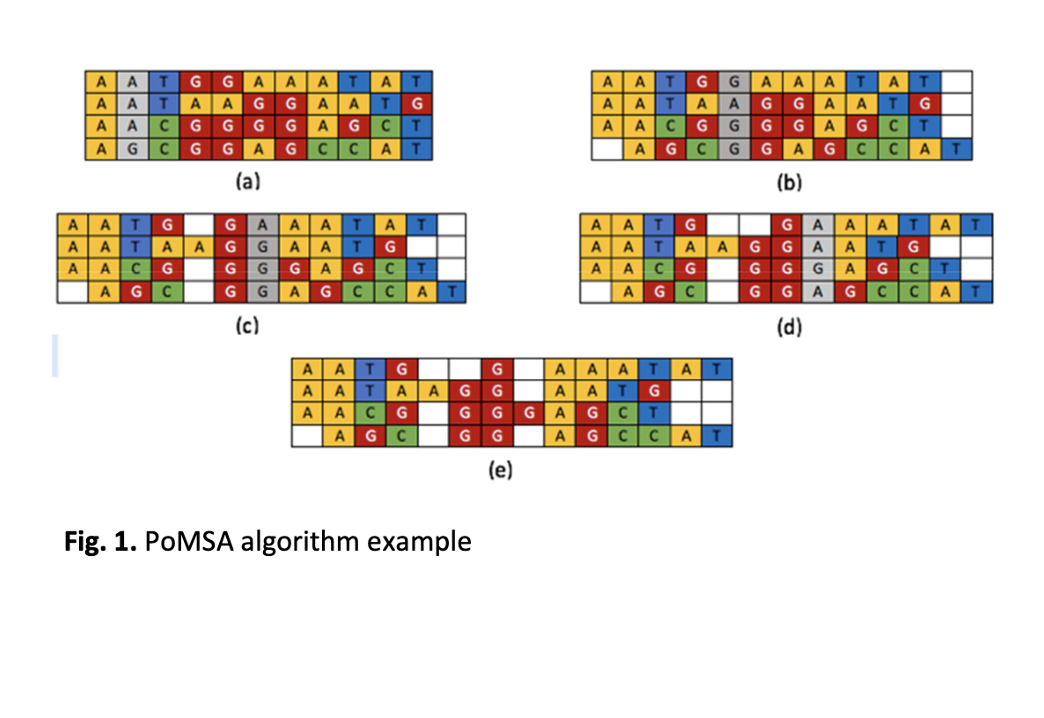
Analyzing the relation between a set of biological sequences can help to identify and understand the evolutionary history of these sequences and the functional relations among them. Multiple Sequence Alignment (MSA) is the main obstacle to proper design and develop homology and evolutionary modeling applications since these kinds of applications require an effective MSA technique with high accuracy. This work proposes a novel Position-based Multiple Sequence Alignment (PoMSA) technique -- which depends on generating a position matrix for a given set of biological sequences. This position matrix can be used to reconstruct the given set of sequences in more aligned format. On the contrary of existing techniques, PoMSA uses position matrix instead of distance matrix to correctly adding gaps in sequences which improve the efficiency of the alignment operation. We have evaluated the proposed technique with different datasets benchmarks such as BAliBASE, OXBench, and SMART. The experiments show that PoMSA technique satisfies higher alignment score compared to existing state-of-art algorithms: Clustal-Omega, MAFTT, and MUSCLE.
Downloads